Introduction to the Entrez search system at NCBI
As you may have read already, NCBI is a public institution that holds information about biological sequences. It’s principal internet page is located at http://www.ncbi.nlm.nih.gov/ . Such webpage connects to all services available at NCBI (PubMed, Entrez, Blast, OMIM, Taxonomy, Structure) as the dark-blue bar indicates in the next figure 1, extracted from their homepage. Although Entrez is indicated as only being one of the offered services, in reality almost all other services are dependent of the database behind the Entrez query system (BLAST needs such databases as well). Entrez is a interface that allows us to explore the contents of the Genbank databases.

Figure 1
Let’s begin a quick tour through the databases. First, let’s visit PubMed. This database contains information about scientific publications and the records have been compiled by the NLM (National Library of Medicine) as an independent effort from Genbank itself, with the colaboration of many publishing companies. There you will find most, in not all of any reference (citations) that you may look for (back to sometime in the 60’s), and many of these records include an abstract. To obtain help about how to make queries, please go to this URL: http://www.ncbi.nlm.nih.gov/entrez/query/static/help/pmhelp.html
Figure 1 shows a menu of databases (currently “GenBank” is selected for searching). The query commands must be entered in the whitespace next to “for”. A query must have a format like this:
"searchword"[field] <logic operator> "searchword"[field] …
Where search-word is the word that identifies a record in the specified “field”. For example, a search word (or search “string”) could be “Johnson” to be searched in the “author” field. Logic operator could be any of the standard “Boolean” operators (AND, OR, NOT). When you write the query command, do not use the “<>” . These were written there only as an idicator. Aditionally, the use of quote signs is optional, but recommended. Boolean operators should be in UPPERCASE.
For example, if you wish to find all papers in 1999 by “Johnson et al” in the “Science” journal, you should use a command like this:
“Johnson”[AU] AND 1999[DP] AND “science”[TA]
Where “AU” is the author field, “DP” is the Date of Publication field and “TA” is the Journal title field. The more information you provide regarding the specific papers you are looking for, the narrower the returned results will be; for instance, if you include more author names.
The most common fields in PubMed that you can use to refine your query are these (pasted from NCBI’s entrez help page):
- All Fields [ALL] Includes all searchable PubMed fields.
- Author Name [AU] Various limits on the number of author names included in the MEDLINE citation have existed over the years (see NLM policy on author names). MEDLINE does not list the full name. The format to search for an author name is: last name followed by a space and up to the first two initials followed by a space and a suffix abbreviation, if applicable, all without periods or a comma after the last name (e.g., fauci as or o'brien jc jr). Initials and suffixes may be omitted when searching. PubMed automatically truncates on an author's name to account for varying initials, e.g., o'brien j [au] will retrieve o'brien ja, o'brien jb, o'brien jc jr, as well as o'brien j. To turn off this automatic truncation, enclose the author's name in double quotes and qualify with [au] in brackets, e.g., "o'brien j" [au] to retrieve just o'brien j.
- EC/RN Number [RN] Number assigned by the Enzyme Commission to designate a particular enzyme or by the Chemical Abstracts Service (CAS) for Registry Numbers.
- Entrez Date [EDAT] Date the citation was added to the PubMed database. Citations are displayed in Entrez Date order which is last in, first out. Dates or date ranges must be entered using the format YYYY/MM/DD [edat], e.g. 1998/04/06 [edat] . The month and day are optional (e.g., 1998 [edat] or 1998/03 [edat]). To enter a date range, insert a colon (:) between each date (e.g., 1996:1997 [edat] or 1998/01:1998/04 [edat]).
- Issue [IP] The number of the journal issue in which the article is published.
- Journal Title [TA] The journal title abbreviation, full journal name, or ISSN number.
- Language [LA]
- Publication Date [DP] The date that the article was published. Dates or date ranges must be searched using the format YYYY/MM/DD [dp], e.g. 1998/03/06 [dp] . The month and day are optional (e.g., 1998 [dp] or 1998/03 [dp]). To enter a date range, insert a colon (:) between each date (e.g., 1996:1998 [dp] or 1998/01:1998/04 [dp]).
- Substance Name [NM] The name of a chemical discussed in the article. Synonyms to the Supplementary Concept Substance Name will automatically map when qualified with [nm]. This field was implemented in mid-1980. Many chemical names are searchable as MeSH terms before that date.
- Text Words [TW] Includes all words and numbers in the title and abstract, and MeSH terms, subheadings, chemical substance names, personal name as subject, and MEDLINE Secondary Source (SI) field. The Personal Name of Subject field can also be searched directly using the search field tag [ps], e.g., nightingale f [ps].
- Title Words [TI] Words and numbers included in the title of a citation.
- Title/Abstract Words [TIAB] Words and numbers included in the title and abstract of a citation.
- Unique Identifiers [UID] PubMed Unique Identifier PMID and MEDLINE Unique Identifier UI .
- Volume [VI] The number of the journal volume in which an article is published.
_______________
Now let’s visit the ENTREZ page. You can click on “Entrez” in the menu bar of NCBI’s homepage as shown in the first figure.
Entrez is a search system that locates/retrieves biological sequence information in the Genbank database. There are several interfaces, and we will concentrate in the web interface. This web interface has the protein and nucleic acid data, the tridimensional structures of some proteins and the full genomes in separate places.
You can make sophisticated queries in order to obtain a set of sequences of interest, for example, you can request all genomic sequences of Arabidopsis that were included in the database from 1997 to 1999 that also contain annotations (in the “feature table”) about promotor regions.
Figure 2 shows the entry page of Entrez for searching the “Nucleotide” database.
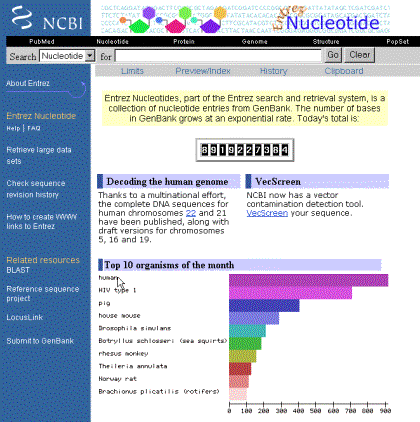
Figure2.
If you pay attention to the frame in the left side of figure 2, there is a submenu with links to different “flavors” of the web-entrez query system. One of the most interesting is that which allows us to obtain a large number of sequences with one single question or query (if you click on “retrieve large datasets”). Another one (“Submit to Genbank”) is used to send sequences of yours to the NCBI so that they get included in the database. This is a more complex process, and is not the scope of this text, but it is useful to know where it is for future references.
In the “Search” whitespace, you can query for sequences using identifying numbers or “index numbers” (such as the gi-number or the accession number). You can also make more complicated queries using the same syntaxis that you used for PubMed.
"searchword"[field] <logic operator> "searchword"[field] …
The most commonly used fields under Entrez are the following:
|
Field |
Definition |
Qualifier |
|
Accession |
Contains the unique accession number of the sequence
or record, assigned to the nucleotide, protein, structure, genome record, or
PopSet by a sequence database builder. The Structure database accession index
contains the PDB IDs but not the MMDB IDs. |
[ACCN] |
|
All Fields |
Contains all terms from all searchable database fields
in the database. |
[ALL] |
|
Author Name |
Contains all authors from all references in the
database records. The format is last name space first initial(s), without
punctuation (e.g., marley jf). |
[AUTH] |
|
EC/RN
Number |
Number assigned by the Enzyme Commission or Chemical
Abstract Service (CAS) to designate a particular enzyme or chemical,
respectively. |
[ECNO] |
|
Feature Key |
Contains the biological features assigned or annotated
to the nucleotide sequences and defined in the DDBJ/EMBL/GenBank Feature
Table (http://www.ncbi.nlm.nih.gov/collab/FT/index.html). Not available for
the Protein or Structure databases. |
[FKEY] |
|
Gene Name |
Contains the standard and common names of genes found
in the database records. This field is not available in Structure database. |
[GENE] |
|
Issue |
Contains the issue number of the journal in which the
data were published. |
[ISS] |
|
Journal Name |
Contains the name of the journal in which the data
were published. Journal names are indexed in the database in abbreviated form
(e.g., J Biol Chem). Journals are also indexed by their by ISSNs. Browse the
index if you do not know the ISSN or are not sure how a particular journal
name is abbreviated. |
[JOUR] |
|
Keyword |
Contains special index terms from the controlled
vocabularies associated with the GenBank, EMBL, DDBJ, SWISS-Prot, PIR, PRF, or
PDB databases. Browse the Keyword indexes of the individual databases to
become familiar with these vocabularies. A Keyword index is not available in
the Structure database. |
[KYWD] |
|
Modification
Date |
Contains the date that records are added to GenBank,
in the format YYYY/MM/DD (e.g., 1999/08/05). A year alone, (e.g., 1984) will
retrieve all records for that year; a year and month (e.g., 1984/03) will
retrieve all records for that month. |
[MDAT] |
|
Organism |
Contains the scientific and common names for the
organisms associated with protein and nucleotide sequences. |
[ORGN] |
|
Page Number |
Contains the number of the first journal page of the
article in which the data were published. |
[PAGE] |
|
Primary
Accession |
Contains the primary accession number of the sequence
or record, assigned to the nucleotide, protein, structure, genome record, or
PopSet by a sequence database builder. A Primary Accession index is not
available in the Structure database. |
[PACC] |
|
Properties |
Contains properties of the nucleotide or protein
sequence. For example, the Nucleotide database's Properties index includes
molecule types, publication status, molecule locations, and GenBank
divisions. A Properties index is not available in the Structure database. |
[PROP] |
|
Protein
Name |
Contains the standard names of proteins found in
database records. Common names may not be indexed in this field so it is best
to also consider All Fields or Text Words. A Protein Name index is not available
in the Structure database. |
[PROT] |
|
Publication
Date |
Contains the date that the record was added to
GenBank, in the format YYYY/MM/DD (e.g., 1999/08/05). A year alone, (e.g.,
1984) will retrieve all records for that year; a year and month (e.g.
1984/03) will retrieve all records for that month. |
[PDAT] |
|
SeqID
String |
Contains the special string identifier, similar to a
FASTA identifier, for a given sequence. A SeqID String index is not available
in the Structure database. |
[SQID] |
|
Sequence
Length |
Contains the total length of the sequence. Sequence
Length indexes are not available in the Structure or PopSet databases. |
[SLEN] |
|
Substance
Name |
Contains the names of any chemicals associated with
this record from the CAS registry and the MEDLINE Name of Substance field.
Substance Name indexes are not available in the Genome or PopSet databases. |
[SUBS] |
|
Text Word |
Contains all of the "free text" associated
with a record. |
[WORD] |
|
Title Word |
Includes only those words found in the definition line
of a record. The definition line summarizes the biology of the sequence and
is carefully constructed by database staff. A standard definition line will
include the organism, product name, gene symbol, molecule type and whether it
is a partial or complete cds. Title Word indexes are not available in the
Structure or PopSet databases. |
[TITL] |
|
Uid |
Contains the Medline unique identifier for records
that contain published references that are linked to PubMed. The Uid index is
not browsable. |
[UID] |
|
Volume |
Contains the volume number of the journal in which the
data were published. |
[VOL] |
Now let’s visit the Taxonomy database:
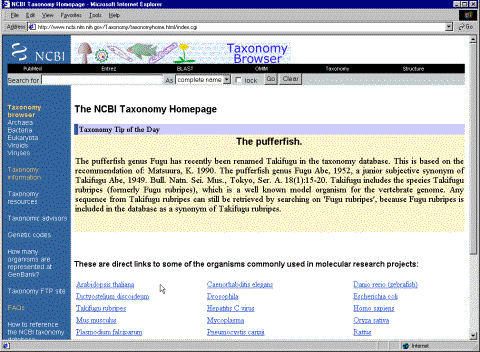
Figure 3.
This page contains the “Tree of Life”, the taxonomic classification is based on data gathered by many systematic biologists that colaborate with the database. It serves as a bridge for associating between sequences in Genbank and the hierarchy of the species origin of the sequences retreived. If we click on “taxonomy browser, eukaryota”, a new page is displayed with a summary of its classification and that allows to check how mnay biological sequences in the database originate from every subdivisions of this taxon (Eukaryota). The number in red is the number of nucleotide sequences for a particular group and the number in blue is the protein sequences. You can also check on “Structures” and “genomes” to display a count of them as well.
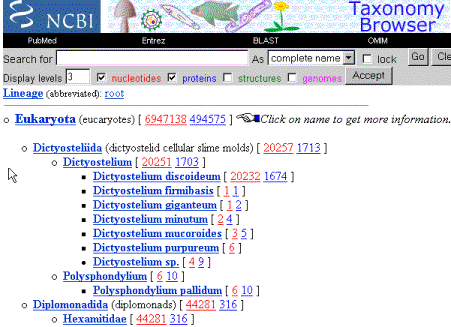
If you wanted to observe the “seed plant” taxon, you would ask for the “spermatophytas” classification:

I f you kept on exploring this database, you would learn what taxonomic name you should use when you want to filter BLAST reports after performing a BLAST search against Genbank at the NCBI webserver. For instance, if you wanted to search your unkown sequence against the “non-redundant” nucleotide database (see the “Sequence comparisons and scores” chapter of these webpages), but filtering so that the program reports are limited to the flowering plants, instead of all of the “nr” database (i.e. not interested in animals and other sequences), you should specify as a filter the taxonomic group “magnoliophyta”.
In the previous figure you can already see that the flowering plants have as of today 749,698 DNA sequences.
=========
Aditional required reading (as indicated at the beggining of this page):
Please read chapter 5 “Information Retrieval from Biological Databases” . Starting at page p.98 of Baxevanis et al. book. Read the introduction but you do NOT need to read pages p.99-100, which deal with the e-mail version of Entrez. Instead skip to page p.101 containing “Integrated Information Retrieval: The Entrez system” heading and keep reading until the end of the chapter.
AND:
We suggest that you review and read this internet page, that contains the help for using entrez:
http://www.ncbi.nlm.nih.gov/entrez/query/static/help/helpdoc.html
=========