EXTERNAL TUTORIALS
Next we will take a tour
through the NCBI pages, specifically, we will read the pages that teach
how to use the Entrez query system and the family of BLAST programs in their
webserver. Please maintain this window open so that it is easy for you
to come back everytime you are done with each of the external links.
1. Entrez
Read the Introduction to the Entrez search system at NCBI if you haven't done this already (it was part of the introductory readings, and you need to know this for item 3 below). After reading it, continue with the following tutorial.
1.
Basic
entrez tutorial. This is an interactive tutorial at the NCBI,
where you will start by searching the nucleotide database with a simplified searching form (no need to explicitly write in boolean commands). For power searches though, the recommended way is to directly search the database with the already explained commands.
2. Problem set.
There is a list of questions and answers to “exercises” with refined
entrez searches that you could use to test yourself in your entrez-searching
capabilites. If you read the required book chapters, you should know by now what
ASN.1 notation means. Some of the questions in the problem set may involve dealing with a page containing ASN1 code.
3. Dowloading in Bulk. If you are
interested in downloading a large set of sequences using the web interface,
you can do this with Batch entrez. A link is available from the normal entrez
page, it is in the blue box on the left side of the page and it reads
“Batch entrez: Retrieve large data sets”.
Read this for help.
Keep in mind that there is a limit (currently 30 minutes) for your
connection to the batch-entrez server, so once you figure what you want to
retrieve (the formulated query), ask to download the gi-numbers first,
then you need to split up your gi-number list into smaller sets so that each
fit the time limit for download, and request each of them by separate. I usually have success with 15,000 EST
sequences per time slot: If I have a
query that will retrieve many sequences (like all Oryza ESTs), I download the list of Gi-numbers using the
same query in batch-entrez, then I use a word processor to break up the list.
For instance, I tell the word processor to find line number 15,000 and I break
the list there, save the new list to a separate text file, then do the same
with the next 15,000.
Because sequences have a wide
range of sizes, you should try a number that fits both the speed of your conection
to the internet (for instance, if using a modem, then request smaller batches
of sequences) and the type of sequences about to be downloaded. If you are
downloading 15,000 full-BAC sequences, is going to take much longer that the
same amount of ESTs, right?
2. Database scanning
In database searches, the query sequence is compared with each of the members in the database using local alignment. The problem with full “ dynamic programming” algorithms such as the Smith&Waterman algorithm (or S&W) is that they are slow (or computationally expensive) for very large sequences or for large databases because they explore the entire space of possible alignments in each pairwise comparison. A database such as dbEST release 010700 (January 2000) containing 3,458,198 sequences had around 1,320,000,000 nucleotide bases (a rounded estimate). The comparison of a single sequence of length 300bp against this database requires creating a matrix with 3.96x11 cells (equal to the product of the sequence length and the database length).This can easily take several hours to compute in a standard modern workstation, making the optimal dynamic algorithm methods impractical.
Because of this, programs that use heuristics have been written, and shown to be much faster than Smith-Waterman (getting the result in seconds or minutes). They were designed to reduce the number of alignments pursued per pairwise comparison while still obtaining the high scoring ones. These programs, by “cutting some corners” can be tens of times faster but run the risk of missing some important true alignments. Among the many existing programs of database searching two became widely accepted, FASTA and BLAST.
FASTA was the first one to be introduced, and has been gradually replaced by BLAST because of better speed and statistics. Until the second version of BLAST arrived, FASTA, even though it was a little slower, was considered better (even up to these days, FASTA has followers who prefer it to BLAST). The first version of BLAST had an excellent statistical analysis of the validity of the results, but it had the disadvantage of not being able to allow gaps in the alignments, so what would be represented as a single alignment in S&W and FASTA programs, if it had any gaps, oldBLAST would represent it as a set of separate alignments. The second version of BLAST has some improvements that made it more sensitive and faster besides allowing for gaps. The acronym BLAST stands for Basic Local Alignment Tool.
The FASTA program is faster than full dynamic programming algorithms (like S&W) because it "cuts around the corners" (uses an exclusion method). Essentially, the core of FASTA first performs a "look up" of common substrings (words or patterns) between the query sequence and the database to identify potential word matches. It does it like this: it divides the query sequence in all possible words of size "K", a parameter that you as the user define, with a default value of six (these words are called Ktups, such as in triplets for k=3), then it looks up that new table of Ktups agaisnt the database and identifies regions in the database (sequences) that match the words. It concentrates later in those sequences that have several nearby matches to the word list; specially if the nearby matches are consecutive.
This is very similar to doing a "find" in your favorite word processor, in which you search for a specific pattern against all the text. In your word processor the "find" little program will show you which lines or paragraphs contain your list of words or phrase, the words being next to each other. It does this by looking up the words that compose your query phrase against the text. The algorithm that does this word search in FASTA is a similar one (not based on alignment) and is many times faster than attempting to align a full paragraph (the analog of a query sequence) against all of the text in the word processor.
So, once the FASTA program has a list of sequence candidates, it uses a dynamic programming algorithm alignment between the query and the identified subset of the database (similar to Smith & Waterman), but limiting the alignment algorithm to a "band" around the region (banded S&W) where the original consecutive word matches were found. This step is much faster than performing full alignments of the query sequence with every other sequence in the database (many of those are unrelated anyway). See picture 1 for a graphical definition of FASTA. Since the introduction of BLAST in 1990, FASTA has also evolved, adquiring some of the good ideas from the contender (such as better statistics reporting). However, it seems that BLAST continues to be the dominant database scanner, in terms of popularity.
The BLAST programs (old and new) gain speed by first creating an index of all possible substrings (words) of length "Word-size" in the query string (with a default of 11 letters for nucleotides and 3 for amino acids) and does a word pattern matching (or look up) of the list against the database. So far, this is similar to what FASTA does, but uses a different word matching algorithm, which is even faster than FASTA's: BLAST adds a little trick to the word pattern matching. To the existing list of possible words derived from the query, it adds words that are the "conserved" variations of each of the originals, allowing the algorithm to find imperfect matches (containing some mismatches) by doing a fast lookup of this expanded list against the database. This is specially practical when performing protein comparisons, since it is easier to expand a list with "conservative" aminoacid variants.
For every pair of sequences (query and target) that have a word in common, the program starts (or "extends") an alignment in both directions of the matching word to determine whether an MSP with score above the threshold has been found. This is several degrees of magnitude faster than starting an alignment of the query sequence against every element in the database. See picture 2 for a graphical definition of the first versions of BLAST.
BLAST 2.0 and above have an additional exclusion step: It only triggers an extension of an alignment between the query and a matched sequence when two (instead of only one) matching words are found in the same diagonal of alignment, and they are within a window of a certain number of base pairs (20 bases is the default). This strategy helped eliminate many random word matches that fire up an alignment that would probably fail to score above the threshold. In BLAST, the slower portion of the program is the extension of word hits into local alignments, based on dynamic programming (the one part that resembles S&W), so any significant cuts of failed extensions is a big jump in speed. Note that dynamic programming used in BLAST2.0 is not limited to a "band" near the original diagonal that fired up the extension (like in FASTA), but it is not the full dynamic programming algorithm either.
Using BLAST
BLAST is actually a set of five programs instead of a single one. These are blastp, blastn, blastx, tblastn and tblastx. Everybody refers to them collectivelly as BLAST, and they all have the same purpose of aligning biological sequences. The difference is on the type of sequences being aligned. For instance:
blastn compares a nucleotide query sequence against a nucleotide sequence database;
blastp compares an amino acid query sequence against a protein sequence database;
blastx compares the six-frame conceptual translation products of a nucleotide query sequence (both strands) against a protein sequence database. This translation is the simple conversion of a nucleotide string into six separate strings of aminoacids (one for each possible reading frame), and is done only once (assuming only one sequence is on the query).
tblastn compares a protein query sequence against a nucleotide sequence database dinamically translated in all six reading frames (both strands). Dynamically on this context means "on the fly": as the program is doing pairwise comparisons between the protein query and the target sequences in the nucleotide database, it is simultaneously translating each target into six posible proteins, all this just before doing the alignments and prior to dealing with the next target in the database.
tblastx compares the six-frame translation of a nucleotide query sequence against the six- frame dynamic translations of a nucleotide sequence database. As you can imagine, this program is doing 36 comparisons (6x6) for each comparison between the query sequence and any of the target sequences in the database. This will of course reflect on the speed of the program, making this one the slowest of the pack. However, this simultaneous translation into protein of both the query (nucleotide) and the target database (also nucleotide), allows us to find more distantly related sequences. When two homologous genes have diverged sufficiently (and accumulated differences) as to make it difficult to find a good and strong alignment, the proteins that both genes code for are more likelly to have conserved sequence similarity if the function of both genes is still the same.
From among the whole set of programs in the BLAST2 suit, the tblastx program is the only program that is unable to perform gapped alignments. This means that it behaves just as the old BLAST set of programs did, placing continous alignments that contain a gap into separate HSPs (separate hits).
The BLAST set of programs have been adapted to several modes of operation. In a local mode of operation, the user downloads the programs to run directly in her/his own computer and downloads databases or creates new ones (with local data, for example). Depending on the size of the databases to search, this might be a very desirable option, but with large databases, it quickly runs out of “machine-power”. A larger workstation, or server machine would be required in that case to run the programs locally.
The second modality is the “network BLAST” in which the user downloads a smaller program that interacts through a network connection with servers running the BLAST programs at NCBI (remotely). The little program that the user downloads is called a “client” and it interfaces with the “server” that runs the heavy load of the job.
The last and more common of the BLAST modalities is the “web BLAST“. This is a simplification of the network blast in the sense that the programs also run remotely inside of servers at the NCBI, but the user does not need to download any client program (or interface). Instead, it uses the existing web capabilities of many computers, running in a web browser such as Netscape or Explorer.
For the course, you are only required to understand and to be able to run the web version of BLAST. Here are some pages that will help you get started. We assume that you already read the first section of reading materials (including first portion of chapter 7 of Baxevanis's book: pages 145-150).
Required Book Reading:
Pages 156-165.
Please read about pairwise (both global and local) alignment in the following sections from chapter 7 (page 145) from Baxevanis et al Book: Start at page 156 and read "database similarity searching", "FASTA", "BLAST", "using BLAST", "recent improvements to BLAST" and stop at the end of page 165.
BLAST tutorial for newbies. Just be careful with the ugly "percent homology" and remember that they probably wanted to say "percent similarity".
When you are done with the
basic tutorial close that window and go ahead and start another one with the next level tutorial of BLAST. This time follow the links in the table at
the end (a squared area with a table of contents, click on the images). The
very first one link in that table is the tutorial you just read, so skip it and
continue with the second one that deals with parameters and interpreting
results. After you finish this one, visit the third one. Here you will
read more advanced information about the interpretation of BLAST output. Then skip to the fifth link page: see
the “Rules of Thumb page” (you will see the link once inside). Close the window
and return to this page. You don’t need
to read anything related to PSI-BLAST or PHI-BLAST at this point.
You will notice that the web
version of BLAST has limited set of parameters to modify. Well, BLAST has MANY parameters that you can
modify to suit your needs, and the default values in the web are’nt necessarily
the best ones (al though they are the fastest ones). The advanced web version of BLAST allows you to modify other
parameters by typing them in a special box.
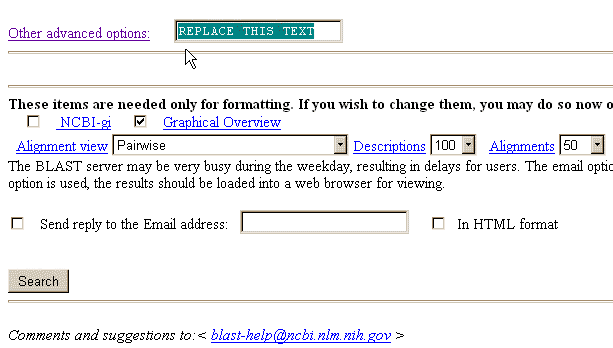
Simply go the the BLAST webpage, choose the "advanced blast" and scroll to the end
of the page. There is a box to fill in
after the text “Other advanced options…”
See next figure:
A full list of parameters is
available in this text file
(which is actually the output of the standalone BLAST program).
Coffee Break at NCBI. See how BLAST is used for real scientific discoveries. Read the introduction if you want and move to the "Contents" using the link in the top bar. Here you can read the latest of the stories, or you can browse the "archives"(bottom bar). Navigate the links to the right of the introductory text of the stories. One such links is a mini "blast tutorial" specifically using the example related to the story that you are reading at the moment. Feel free to roam around these pages; they contain very inspiring examples of how to use sequence alignment. Pay particular attention at what parameters were used (whenever they specify them) and why they were more useful this way. If you have the time read all of these stories.
The
best examples (personal opinion) are these:
![]() Horizontal
transfer. How some plant genes made
their way into a human parasite.
Horizontal
transfer. How some plant genes made
their way into a human parasite.
![]() Analysis
of the developmental program in the compound eye of the fly (and lessons to
learn from it).
Analysis
of the developmental program in the compound eye of the fly (and lessons to
learn from it).
![]() How
does the cell detect abnormal mRNAs and defends itself from truncated proteins.
How
does the cell detect abnormal mRNAs and defends itself from truncated proteins.
Now we will
leave the NCBI servers and take other tours:
Using FASTA
The original FASTA service is at the University of Virginia where you can use the program to search against the standard NCBI databases. The EBI also offers a webservice (EMBL outstation) where you can run a FASTA search against the EMBL databases. You can access this service in this page: http://www2.ebi.ac.uk/fasta3/?request
I have not been able to find an interactive tutorial on the FASTA programs like the one for BLAST at the NCBI, but if you read the FASTA help page at the EMBL outstation, and have finished the BLAST online tutorial, performing a FASTA search should not be difficult, since is analogous to BLAST (The webservices' interfaces are similar). In essence, you have to choose a database to search, a program flavor(fasta3, fastx3, tfastx3, fasts3, fastf3 and others) and some parameters for the execution of the program.
3. Multiple alignment with CLUSTALX
CLUSTALX is graphical interface to the otherwise "tedious" command line program CLUSTALW. This program is the most commonly used when aligning multiple sequences. There are serveral free versions of this program, both for downloading and for running remotely on servers. Because it is so common, it has also been implemented in other commercial bioinformatics program suites (like SeqLab (GCG) and DNASTAR lasergene).
Required Book Reading:
Pages 172-188.
Please read chapter 8, that deals with multiple alignment with programs such as ClustalW and also with exploring patterns and motifs in groups of aligned sequences. The chapter starts at page 172 and finishes on page 188.
There is is a tutorial on how to do multiple sequence alignment (but with a very advanced level)here.
For running CLUSTALX using a web interface, you can use the following link:
The BCM launcher also has a web interface to CLUSTALX. Or you can choose a variety of algorithms different from CLUSTALX:
Here is a list of extra (optional) useful links: http://ascus.plbr.cornell.edu/PB607/Useful-links.html. ![]() ClustalW at the EMBL Outstation. http://www.ebi.ac.uk/clustalw/
ClustalW at the EMBL Outstation. http://www.ebi.ac.uk/clustalw/
![]() http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html
http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html
![]()