 dziej *,
dziej *, 
 mierkiewicz *,
mierkiewicz *, 
BIOPHYSICS
Physics-based protein-structure prediction using a
hierarchical protocol based on the UNRES force field: Assessment in two blind
tests
 dziej *,
dziej *, 
 mierkiewicz *,
mierkiewicz *, 
*Baker Laboratory of Chemistry and Chemical Biology, Cornell
University, Ithaca, NY 14853-1301;
 Faculty of Chemistry,
University of Gda
Faculty of Chemistry,
University of Gda sk, Sobieskiego Str.
18, 80-952 Gda
sk, Sobieskiego Str.
18, 80-952 Gda sk, Poland;
sk, Poland;
 Instituto de Matemática
Aplicada San Luis, Consejo Nacional de Investigaciones Científicas y Técnicas de
Argentina, Facultad de Ciencias Físico Matemáticas y Naturales, Universidad
Nacional de San Luis, Ejército de los Andes 950, 5700 San Luis, Argentina;
Instituto de Matemática
Aplicada San Luis, Consejo Nacional de Investigaciones Científicas y Técnicas de
Argentina, Facultad de Ciencias Físico Matemáticas y Naturales, Universidad
Nacional de San Luis, Ejército de los Andes 950, 5700 San Luis, Argentina;
 Cornell Theory Center, Cornell
University, Ithaca, NY 14853-3801; and ¶Department of Chemistry,
Chungbuk National University, Cheongju, Chungbuk 361-763, Korea
Cornell Theory Center, Cornell
University, Ithaca, NY 14853-3801; and ¶Department of Chemistry,
Chungbuk National University, Cheongju, Chungbuk 361-763, Korea
Contributed by H. A. Scheraga, March 31, 2005
Recent improvements in the protein-structure prediction method
developed in our laboratory, based on the thermodynamic hypothesis,
are described. The conformational space is searched extensively at
the united-residue level by using our physics-based UNRES energy
function and the conformational space annealing method of global
optimization. The lowest-energy coarse-grained structures are then
converted to an all-atom representation and energy-minimized with the
ECEPP/3 force field. The procedure was assessed in two recent blind
tests of protein-structure prediction. During the first blind test,
we predicted large fragments of  and
and  +
+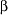 proteins [60–70 residues with C
proteins [60–70 residues with C rms deviation (rmsd) <6 Å]. However, for
rms deviation (rmsd) <6 Å]. However, for
 +
+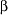 proteins, significant topological errors occurred despite low rmsd
values. In the second exercise, we predicted whole structures of five
proteins (two
proteins, significant topological errors occurred despite low rmsd
values. In the second exercise, we predicted whole structures of five
proteins (two  and three
and three
 +
+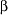 ,
with sizes of 53–235 residues) with remarkably good accuracy. In
particular, for the genomic target TM0487 (a 102-residue
,
with sizes of 53–235 residues) with remarkably good accuracy. In
particular, for the genomic target TM0487 (a 102-residue
 +
+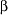 protein from Thermotoga maritima), we predicted the complete,
topologically correct structure with 7.3-Å C
protein from Thermotoga maritima), we predicted the complete,
topologically correct structure with 7.3-Å C rmsd. So far this protein is the largest
rmsd. So far this protein is the largest
 +
+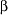 protein predicted based solely on the amino acid sequence and a
physics-based potential-energy function and search procedure. For
target T0198, a phosphate transport system regulator PhoU from T.
maritima (a 235-residue mainly
protein predicted based solely on the amino acid sequence and a
physics-based potential-energy function and search procedure. For
target T0198, a phosphate transport system regulator PhoU from T.
maritima (a 235-residue mainly
 -helical protein), we predicted
the topology of the whole six-helix bundle correctly within 8 Å rmsd,
except the 32 C-terminal residues, most of which form a
-helical protein), we predicted
the topology of the whole six-helix bundle correctly within 8 Å rmsd,
except the 32 C-terminal residues, most of which form a
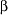 -hairpin. These and other
examples described in this work demonstrate significant progress in
physics-based protein-structure prediction.
-hairpin. These and other
examples described in this work demonstrate significant progress in
physics-based protein-structure prediction.